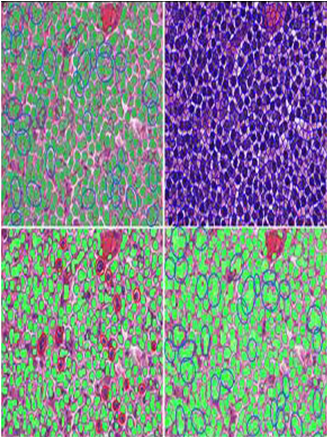
Cell Nuclei Segmentation
This approach for the detection of cells from H&E-stained tissues, involves an optimal color space, called MDC. Mathematically, we aim to construct the new MDC color space by where I* = [Im, Id, Ic] represents the MDC color space transformed pixel, I = [Ir, Ig, Ib] represents the RGB space, A is a 3x3 coefficient matrix for constructing MDC from the RGB color space. In order to account for regional differences due to staining, the A matrix will be calculated in each locale; hence, it will be adaptive. In our experience, the best representation of microscopic images is by means of color texture features such as Local Fourier Transforms. The figure shows a representative segmentation result along with a comparison to other commonly used techniques.
The PIIP API will provide the input images necessary for the region of interest where the nuclei segmentation algorithm needs to be implemented. If the selected image is larger than the memory limits of the running system, the PIIP will automatically divide up the image into appropriate regions and process each region separately and then combine the outputs. For efficiency purposes, each of the regions may be run on a separate processor. The output can be displayed in several forms: segmentation masks of the cells, cell boundaries (by a line around the nuclei), or a single point in the detected cell centroid. All of this information will be automatically generated by PIIP. By using these sets of outputs, the algorithmic evaluation module can be run efficiently. Additionally, the PIIP viewer will be able to display these results to the user at a single location or multiple locations. As some users may want to see algorithmic outputs in different ways (e.g. single centroid and cell boundaries), the PIIP will provide multiple ways of displaying output.